bssm: Bayesian Inference of Non-linear and Non-Gaussian State Space Models in R
Jouni Helske (joint work with Matti Vihola)
University of Jyväskylä, Finland
9/7/2021
1 / 15
What are state space models?
- The bssm package (Helske, Vihola, 2021) allows fully Bayesian inference of state space models (SSMs)
- E.g. structural time series, ARIMA models, generalized linear models with time-varying coefficients, cubic splines, SDEs, ...
- In general we have
- Observations y=(y1,…,yT) with conditional distribution gt(yt|αt)
- Latent states α=(α1,…,αT) with a transition distribution pt(αt+1|αt)
- Both observations yt and states αt can be multivariate
- Both distributions can depend on some hyperparameters θ
- Our interest is in the posterior distribution p(θ,α|y)
- Prediction problems p(yT+k|y) and interpolation p(yt|y) are also straightforward with SSM setting
2 / 15
But wait, what about KFAS?
- Compared to the KFAS package (Helske, 2017) for state space modelling:
- KFAS is mainly for maximum likelihood inference vs Bayesian approach of bssm
- bssm supports more model types (nonlinear models, stochastic volatility models, SDEs)
- KFAS uses importance sampling for non-Gaussian models, bssm uses particle filtering (scales better)
- bssm is easier to maintain and extend further (written in C++ instead of Fortran)
- Creating models is more user-friendly with KFAS (but see
as_bssmfunction!)
3 / 15
Bayesian estimation of SSMs
- Two important special cases:
- Both observation equation and state equation are linear-Gaussian
- States αt are categorical (often called hidden Markov models, not covered by bssm)
- In these cases the marginal likelihood p(y|θ) can be computed easily
- Marginalization of latent states results in highly efficient Markov chain Monte Carlo (MCMC) algorithms
- Run MCMC targeting the marginal posterior of hyperparameters θ.
- Given samples from p(θ|y), simulate latent states from the smoothing distribution p(α|y,θ).
- θ is often low dimensional so simple adaptive random walk Metropolis works well.
4 / 15
Bayesian inference for general SSMs
- In general, marginal likelihood p(y|θ) is not analytically tractable. Three routes forward:
- Sample both θ and α directly using, e.g., BUGS (Lunn et al. 2000) or Stan (Stan Development Team 2021). Typically inefficient due to strong correlation structures and high dimensionality of α.
- Use (deterministic) approximation of p(y|θ), e.g, INLA (Rue et al. 2009), extended Kalman filter (EKF). Fast(er) but biased. Bias is hard to quantify.
- Use particle MCMC (Andrieu et al. 2010) where p(y|θ) is replaced with its unbiased estimator from particle filter. Leads to asymptotically exact inference, but often computationally intensive. Tuning of MCMC nontrivial with respect to number of particles and acceptance rate.
5 / 15
IS-MCMC for state space models
- What if we could combine fast approximations and exact methods?
- Vihola, Helske and Franks (2020) suggest targeting an approximate marginal posterior of θ, combined with importance sampling type post-correction (IS-MCMC):
- Given θ, assume that we can compute approximation ^p(y|θ)=p(y|θ)/w(θ).
- Run MCMC targeting ^p(θ|y), where the marginal likelihood is replaced with the the approximation ^p(y|θ).
- For each θ from approximate marginal, run particle filter to obtain samples of α and unbiased estimate of p(y|θ).
- We now have weighted samples of (θ,α) from the correct posterior, with weights w(θ)=p(y|θ)/^p(y|θ).
6 / 15
Post-correction
- For post-correction we recommend particle filter called ψ-APF (Vihola, Helske, Franks, 2020), which uses the dynamics of the approximating model with look-ahead strategy.
- Based on the approximating densities ^gt(yt|αt), and ^pt(αt+1|αt)
- Produces correct smoothing distribution and unbiased estimate of the marginal likelihood
For state space models supported by
bssm, often only a small number (e.g. 10) particles is enough for accurate likelihood estimate.Post-correction is easy to parallelize and the needs to be done only for accepted θ.
7 / 15
Linear-Gaussian state space models (LGSSM)
yt=dt+Ztαt+Htϵt,ϵt∼N(0,I)αt+1=ct+Ttαt+Rtηt,ηt∼N(0,I)α1∼N(a1,P1)
- dt, Zt, Ht, ct, Tt, Rt, a1, P1 can depend on θ.
- Kalman filter gives us marginal likelihood p(y|θ).
- Smoothing algorithms give p(α|y,θ).
- Building general LGSSM and some special cases in bssm:
# univariate LGSSM, ssm_mlg for multivariate versionssm_ulg(y, Z, H, T, R, a1, P1, D, C, init_theta, prior_fn, update_fn)# Basic structural time series modelbsm_lg(y, sd_level = gamma(1, 2, 10), sd_y = 1, xreg = X, beta = normal(0, 0, 10))8 / 15
Non-Gaussian observations
- State equation has the same form as in LGSSMs, but observations are non-Gaussian
- For example, gt(yt|αt)=Poisson(utexp(dt+Ztαt)), where ut is the known exposure at time t.
- Filtering, smoothing and likelihood available via sequential Monte Carlo (SMC) i.e. particle filtering.
- Approximate inference possible via Laplace approximation
- Find LGSSM with same mode of p(α|y,θ) (iteratively)
ssm_ung(y, Z, T, R, distribution = "poisson")ssm_mng(...)bsm_ng(...)svm(...)ar1_ng(...)9 / 15
Bivariate Poisson model with bssm
# latent random walkalpha <- cumsum(rnorm(100, sd = 0.1))# observationsy <- cbind(rpois(100, exp(alpha)), rpois(100, exp(alpha)))# function which defines the log-prior densityprior_fun <- function(theta) { dgamma(theta, 2, 0.01, log = TRUE)}# function which returns updated model componentsupdate_fun <- function(theta) { list(R = array(theta, c(1, 1, 1)))}model <- ssm_mng(y = y, Z = matrix(1, 2, 1), T = 1, R = 0.1, P1 = 1, distribution = "poisson", init_theta = 0.1, prior_fn = prior_fun, update_fn = update_fun)10 / 15
Other models supported by bssm
Non-linear Gaussian models: yt=Zt(αt)+Ht(αt)ϵt,αt+1=Tt(αt)+Rt(αt)ηt,α1∼N(a1,P1),
- Unbiased estimation via particle filtering.
- Approximations with mode matching based on extended Kalman filter and smoother.
Models where the state equation is defined as a continuous-time diffusion: dαt=μ(αt)dt+σ(αt)dBt,t≥0,
- Bt is a Brownian motion, μ and σ are real-valued functions
- Observation density pk(yk|αk) defined at integer times k=1…,n.
These use user-defined C++ -snippets for model components based on a template provided
11 / 15
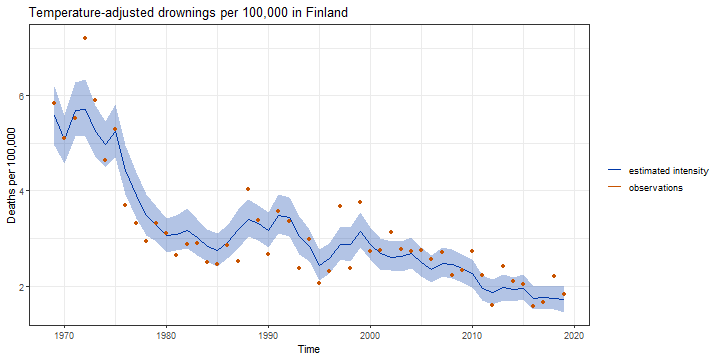
Illustration: Modelling deaths by drowning in Finland 1969-2019
- Yearly drownings yt assumed to follow Poisson distribution
- Predictor xt is (centered) average summer temperature (June to August)
- Exposure ut is the yearly population in hundreds of thousands yt∼Poisson(utexp(βxt+μt))t=1,…,Tμt+1=μt+νt+ηt,ηt∼N(0,σ2η)νt+1=νt+ξt,ξt∼N(0,σ2ξ)
- Hyperparameters θ=(β,ση,σξ)
- Latent states αt=(μt,νt)
12 / 15
Estimating the model with bssm
data("drownings")model <- bsm_ng( y = drownings[, "deaths"], u = drownings[, "population"], xreg = drownings[, "summer_temp"], distribution = "poisson", beta = normal(init = 0, mean = 0, sd = 1), sd_level = gamma(init = 0.1, shape = 2, rate = 10), sd_slope = gamma(0, 2, 10))fit <- run_mcmc(model, iter = 20000, particles = 10)summary(fit, TRUE)[,1:4]## Mean SD SE ESS## sd_level 0.1029887 0.02195574 0.0007928511 766.8547## sd_slope 0.0161829 0.01130342 0.0003732525 917.0944## coef_1 0.1109080 0.01743323 0.0005728851 926.020713 / 15
Decrease in drownings after adjusting temperature and population growth
14 / 15
Thank you!
Some references:
- Helske, J. (2017). KFAS: Exponential Family State Space Models in R. Journal of Statistical Software, 78(10), 1-39. https://www.jstatsoft.org/article/view/v078i10
- Helske J, Vihola M (2021). bssm: Bayesian Inference of Non-linear and Non-Gaussian State Space Models in R. ArXiv preprint 2101.08492, https://arxiv.org/abs/2101.08492
- Vihola M, Helske J, Franks J (2020). Importance Sampling Type Estimators Based on Approximate Marginal MCMC. Scandinavian Journal of Statistics. https://doi.org/10.1111/sjos.12492
- Lunn, D.J., Thomas, A., Best, N., and Spiegelhalter, D. (2000) WinBUGS — a Bayesian modelling framework: concepts, structure, and extensibility. Statistics and Computing, 10:325–337.
- Stan Development Team (2021). Stan Modeling Language Users Guide and Reference Manual, 2.27. https://mc-stan.org
- Rue, H., Martino, S. and Chopin, N. (2009). Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. Journal of the Royal Statistical Society: Series B, 71: 319-392. https://doi.org/10.1111/j.1467-9868.2008.00700.x
- Andrieu, C., Doucet, A. and Holenstein, R. (2010), Particle Markov chain Monte Carlo methods. Journal of the Royal Statistical Society: Series B, 72: 269-342. https://doi.org/10.1111/j.1467-9868.2009.00736.x
15 / 15